Workflow from Scientific Research
CC-BY
3
Views
0
Likes
Citation
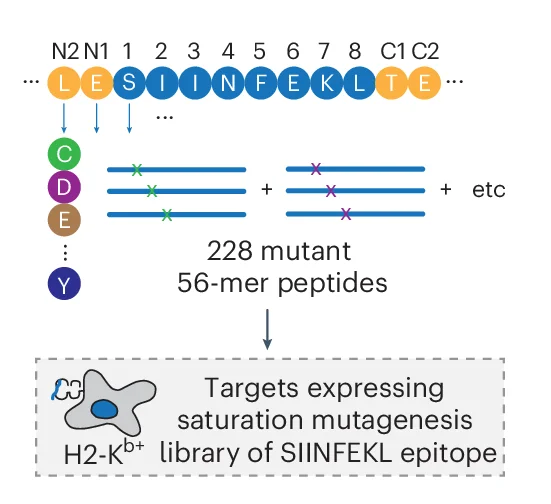
Design of the SIINFEKL epitope saturation mutagenesis library. Each position was substituted to each of the 19 alternative amino acids. In addition to the epitope, the two amino acids that were N terminal and C terminal to the epitope were examined. Each mutant epitope was expressed as 56-aa peptide tiles, introduced into H2-K b + G 5 -target cells and screened using OT-I TCR + SrtA-Jurkats using TCR-MAP.
#Workflow#Illustration#SIINFEKL Epitope#Saturation Mutagenesis Library#Mutant Epitope#Peptide Tiles#H2-K b + G 5 -Target Cells#OT-I TCR#SrtA-Jurkats#TCR-MAP
Related Plots
Browse by Category
Popular Collections
Discover More Scientific Plots
Browse thousands of high-quality scientific visualizations from open-access research